What is the Caviness Cluster and Why Should You Use it?
Overview
Teaching: 25 min
Exercises: 5 minQuestions
What is the Caviness Cluster and who can use it?
What can I expect to learn from this course?
Objectives
Understand the basics about the Caviness Cluster, who are its owners and who can use it.
Identify how the Caviness Cluster could benefit your research.
Know how Caviness may differ from other High Performance Computing (HPC) Systems but also other clusters here at the University of Delaware
What is the Caviness Cluster?
The Caviness cluster is named in honor of Jane Caviness, the former director of Academic Computing Services at the University of Delaware. In the 1980’s, Jane Caviness led a groundbreaking expansion of UD’s computing resources and network infrastructure that laid the foundation for UD’s current research computing capabilities.
Caviness is University of Delaware’s (UD) third community cluster. It was deployed in July 2018 and is a distributed-memory Linux cluster. It is based on a rolling-upgradeable model for expansion and replacement of hardware over time. For more information on the specifics of Caviness hardware please vist the Caviness page. The cluster was purchased with a proposed 5 year life for the first generation hardware, putting its refresh in the April 2023 to June 2023 time period.
Caviness is a technical and financial partnership between UD’s Information Technologies and UD’s faculty and researchers who require high-performance computing resources. Faculty and researchers become financial stakeholders by purchasing HPC resources (based on the cost of one or more compute nodes) in the cluster. Only stakeholders and the researchers sponsored by a stakeholder may use the cluster. It is located at UD’s Computing Center.
Why Use The Caviness Cluster
What do you need?
Talk to your neighbor about your research. How does computing help you do your research? How could more computing help you do more or better research?
Frequently, research problems that use computing can outgrow the desktop or laptop computer where they started:
- A statistics student wants to do cross-validate their model. This involves running the model 1000 times – but each run takes an hour. Running on their laptop will take over a month!
- A genomics researcher has been using small datasets of sequence data, but soon will be receiving a new type of sequencing data that is 10 times as large. It’s already challenging to open the datasets on their computer – analyzing these larger datasets will probably crash it.
- An engineer is using a fluid dynamics package that has an option to run in parallel. So far, they haven’t used this option on their desktop, but in going from 2D to 3D simulations, simulation time has more than tripled and it might be useful to take advantage of that feature.
In all these cases, what is needed is access to more computers that can be used at the same time. Luckily, large scale computing systems – shared computing resources with lots of computers – like Caviness can be use to help with these problems. HPC systems similar to Caviness can be found at many other universities, labs, or through national networks. These resources usually have more central processing units(CPUs), CPUs that operate at higher speeds, more memory, more storage, and faster connections with other computer systems. They are frequently called “clusters”, “supercomputers” or resources for “high performance computing” or HPC. In this lesson, we will usually use the terminology of HPC and HPC cluster.
Using a cluster often has the following advantages for researchers:
- Speed. With many more CPU cores, often with higher performance specs, than a typical laptop or desktop, HPC systems can offer significant speed up.
- Volume. Many HPC systems have both the processing memory (RAM) and disk storage to handle very large amounts of data. Terabytes of RAM and petabytes of storage are available for research projects.
- Efficiency. Many HPC systems operate a pool of resources that are drawn on by a many users. In most cases when the pool is large and diverse enough the resources on the system are used almost constantly.
- Cost. Bulk purchasing and government funding means that the cost to the research community for using these systems is significantly less than it would be otherwise.
- Convenience. Maybe your calculations just take a long time to run or are otherwise inconvenient to run on your personal computer. There’s no need to tie up your own computer for hours when you can use someone else’s instead.
This is how a large-scale compute system like a cluster can help solve problems like those listed at the start of the lesson.
Thinking ahead
How do you think using a large-scale computing system will be different from using your laptop? Talk to your neighbor about some differences you may already know about, and some differences/difficulties you imagine you may run into.
Caviness Cluster compared to the other clusters at UD and other HPC systems.
The workload management software on Caviness is Slurm. It is commonly used for HPC systems but it is new to UD. The prior two community clusters used Grid Engine for their workload manager. For setting up your computing environment Caviness will continue to use VALET, just as Mills and Farber have used. VALET is a recursive acronym for VALET Automates Linux Environment Tasks – is an alternative that strives to be as simple as possible to configure your environment to use software installed on the cluster. VALET is unique to UD, if you have used another HPC system you might be familiar with the Environment Modules Package, more commonly known as modules. VALET and modules essentially perform the same services, but each have a different syntax in doing so. More details on VALET will be coming later on in the course.
On Command Line
Using Caviness like most HPC systems involves the use of a shell through a command line interface (CLI) and either specialized software or programming techniques. The shell is a program with the special role of having the job of running other programs rather than doing calculations or similar tasks itself. Whatever the user types, is passed to the shell, which then figures out what commands to run and orders the computer to execute them. (Note that the shell is called “the shell” because it encloses the operating system in order to hide some of its complexity and make it simpler to interact with.) The most popular Unix shell is Bash, the Bourne Again SHell (so-called because it’s derived from a shell written by Stephen Bourne). Bash is the default shell on most modern implementations of Unix and in most packages that provide Unix-like tools for Windows.
Interacting with the shell is done via a command line interface (CLI) on most HPC systems. In the earliest days of computers, the only way to interact with early computers was to rewire them. From the 1950s to the 1980s most people used line printers. These devices only allowed input and output of the letters, numbers, and punctuation found on a standard keyboard, so programming languages and software interfaces had to be designed around that constraint and text-based interfaces were the way to do this. Typing-based interfaces are often called a command-line interface, or CLI, to distinguish it from a graphical user interface, or GUI, which most people now use. The heart of a CLI is a read-evaluate-print loop, or REPL: when the user types a command and then presses the Enter (or Return) key, the computer reads it, executes it, and prints its output. The user then types another command, and so on until the user logs off.
Learning to use Bash or any other shell sometimes feels more like programming than like using a mouse. Commands are terse (often only a couple of characters long), their names are frequently cryptic, and their output is lines of text rather than something visual like a graph. However, using a command line interface can be extremely powerful, and learning how to use one will allow you to reap the benefits described above.
The rest of this lesson
The only way to use these types of resources is by learning to use the command line. This introduction to Caviness has two parts:
- We will learn to use the UNIX command line (also known as Bash).
- We will use our new Bash skills to connect to and operate a high-performance computing supercomputer.
The skills we learn here have other uses beyond Caviness or other HPC: Bash and UNIX skills are used everywhere, be it for web development, running software, or operating servers. It’s become so essential that Microsoft now ships it as part of Windows! Knowing how to use Bash and HPC systems will allow you to operate virtually any modern device. With all of this in mind, let’s connect to a cluster and get started!
Key Points
Caviness is like other HPC systems but does use a custom program (VALET) for setting up the computing environment.
Caviness and HPC systems can be used to do work that would either be impossible or much slower on a local desktop or lab computer.
The standard method of interacting with most HPC systems is via a command line interface such as Bash.
Connecting to the Caviness Cluster
Overview
Teaching: 20 min
Exercises: 10 minQuestions
How do I open a terminal?
How do I connect to a remote computer?
Objectives
Remotely connect to the Caviness cluster
Opening a Terminal
Connecting to Caviness must be done through a tool known as “SSH” (Secure SHell) and usually it is run through a terminal. So, to begin using Caviness we need to begin by opening a terminal. Different operating systems have different terminals, none of which are exactly the same in terms of their features and abilities while working on the local operating system. When connected to the remote system the experience between terminals will be identical as each will faithfully present the same experience of the Caviness cluster.
Here is the process for opening a terminal in each operating system.
Linux
There are many different versions (aka “flavors”) of Linux and how to open a terminal window can change between flavors. Fortunately, most Linux users already know how to open a terminal window since it is a common part of the workflow for Linux users. If this is something that you do not know how to do then a quick search on the Internet for “how to open a terminal window in” with your particular Linux flavor appended to the end should quickly give you the directions you need.
A very popular version of Linux is Ubuntu. There are many ways to open a terminal window in Ubuntu but a very fast way is to use the terminal shortcut key sequence: Ctrl+Alt+T.
Mac
Macs have had a terminal built in since the first version of OS X since it is built on a UNIX-like operating system, leveraging many parts from BSD (Berkeley Systems Designs). The terminal can be quickly opened through the use of the Searchlight tool. Hold down the command key and press the spacebar. In the search bar that shows up type “terminal”, choose the terminal app from the list of results (it will look like a tiny, black computer screen) and you will be presented with a terminal window. Alternatively, you can find Terminal under “Utilities” in the Applications menu.
XQuartz
XQuartz is an open-source project thats goal is to bring X.org Window Systems to the Mac operating system. It allows for GUI, images, and other graphical interactions on a remote machine to be viewable on a local machine connected to the remote system. For more information about it or to download it visit the XQuartz sites
Windows
While Windows does have a command-line interface known as the “Command Prompt” that has its roots in MS-DOS (Microsoft Disk Operating System) its built-in SSH tool does not have all the options we need in setting up our SSH connections. To get those additional options we will need to install some 3rd party applications. There are a variety of programs that can be used. Below we will go into the details of two more common ones.
PuTTY
It is strictly speaking not necessary to have a terminal running on your local computer in order to access and use a remote system, only a window into the remote system once connected. PuTTy is likely the oldest, most well-known, and widely used software solution to take this approach.
PuTTY is available for free download from UDeploy or www.putty.org. Through the UDeploy site you will have the option to download and install Putty, Xming, and WinSCP. It is suggested that you download and install all three of these programs. The download and installation process will not be covered in this lesson. For directions on how to download, install, and set up PuTTY can be found in the course’s setup lesson.
Running PuTTY will not initially produce a terminal but instead a window full of connection options. Putting the address of the remote system in the “Host Name (or IP Address)” box and either pressing enter or clicking the “Open” button should begin the connection process. If there is a save session you could alternatively “double click” a on the saved session name, or click to highlight it and then click on the “Open” button to start the connection process.
If this works you will see a terminal window open that prompts you for a username through the “login as:” prompt and then for a password. If both of these are passed correctly then you will be given access to the system and will see a message saying so within the terminal. If you need to escape the authentication process you can hold the Control (ctrl) key and press the C key to exit and start again.
Note that you may want to paste in your password rather than typing it. Use ctrl plus a right-click of the mouse or use shift + insert to paste content from the clipboard to the PuTTY terminal.
For those logging in with PuTTY it would likely be best to cover the terminal basics already mentioned above before moving on to navigating the remote system.
XMing
Xming is an X11 display server for Microsoft Windows operating systems, for Windows XP and later. It can be installed to allow a GUI launched from Caviness to display on your Windows computer. This will allow you to interact with a program’s GUI, such as Matlab, or view images of results such as charts or tables. The download and installation process will not be covered in this lesson. For directions on how to download, install, and set up XMing can be found in the course’s setup lesson.
Logging onto the system
With all of this in mind, let’s make a remote connection. In this workshop, we will connect to Caviness — which is one of three community clusters at University of Delaware. Although it’s unlikely that every system will be exactly like Caviness, it’s a very good example of what you can expect from an HPC installation. To connect to our example computer, we will use SSH (if you are using PuTTY, see above).
SSH allows us to connect to UNIX computers remotely, and use them as if they were our own. The
general syntax of the connection command follows the format ssh yourUsername@some.computer.address.
Let’s attempt to connect to the HPC system now:
ssh yourUsername@caviness.hpc.udel.edu
$ ssh traine@caviness.hpc.udel.edu
The authenticity of host 'caviness.hpc.udel.edu (128.4.5.164)' can't be established.
ECDSA key fingerprint is SHA256:bmpB4mr7nmeTgtDHts4yjB73BFMEJAfZ/mSoGgj5jGA.
Are you sure you want to continue connecting (yes/no/[fingerprint])? yes
Warning: Permanently added 'caviness.hpc.udel.edu' (ECDSA) to the list of known hosts.
......................................................................
Caviness cluster (caviness.hpc.udel.edu)
This computer system is maintained by University of Delaware
IT. Links to documentation and other online resources can be
found at:
http://docs.hpc.udel.edu/abstract/caviness/
For support, please contact consult@udel.edu
......................................................................
traine@caviness.hpc.udel.edu's password:
If you’ve connected successfully, you should see a prompt like the one below. This prompt is informative, and lets you grasp certain information at a glance. (If you don’t understand what these things are, don’t worry! We will cover things in depth as we explore the system further.)
[traine@login00 ~]$
Note that Caviness has two login node login00 and login01. The above output could reflect either node.
Telling the Difference between the Local Terminal and the Remote Terminal
You may have noticed that the prompt changed when you logged into the remote system using the
terminal (if you logged in using PuTTY this will not apply because it does not offer a local
terminal). This change is important because it makes it clear on which system the commands you type
will be run when you pass them into the terminal. This change is also a small complication that we
will need to navigate throughout the workshop. Exactly what is reported before the $ in the
terminal when it is connected to the local system and the remote system will typically be different
for every user. We still need to indicate which system we are entering commands on though so we will
adopt the following convention:
[local]$when the command is to be entered on a terminal connected to your local computer[traine@login00 ~]$when the command is to be entered on a terminal connected to the remote system$when it really doesn’t matter which system the terminal is connected to.
Being certain which system your terminal is connected to
If you ever need to be certain which system a terminal you are using is connected to then use the following command:
$ hostname.
Keep two terminal windows open
For Mac and Linux user:
It is strongly recommended that you have two terminals open, one connected to the local system and one connected to the remote system, that you can switch back and forth between. If you only use one terminal window then you will need to reconnect to the remote system using one of the methods above when you see a change from
[local]$to[traine@login00 ~]$and disconnect when you see the reverse.
Key Points
Connect to the Caviness cluster by using SSH:
ssh yourUsername@caviness.hpc.udel.eduConnect to the Caviness cluster by using SSH with X Win (using Xming):
ssh -Y yourUsername@caviness.hpc.udel.edu
Navigating the Caviness Filesystem
Overview
Teaching: 30 min
Exercises: 5 minQuestions
How do I navigate and start interacting with files and directories on Caviness?
How do I keep track of where I am in the filesystem?
Objectives
Learn how to navigate around directories and look at their contents.
Explain the difference between a file and a directory.
Translate an absolute path into a relative path and vice versa.
Identify the actual command, flags, and filenames in a command-line call.
Demonstrate the use of tab completion, and explain its advantages.
At this point in the lesson, we’ve just logged into the system. Nothing has happened yet, and we’re
not going to be able to do anything until we learn a few basic commands. In this lesson we will talk
about ls, cd, and a few other commands. These commands will help you navigate around not only the
Caviness filesystem structure but nearly any Linux/Unix filesystem using the CLI.
Right now, all we see is something that looks like this:
[traine@login00 ~]$
The dollar sign is a prompt, which shows us that the shell is waiting for input. If you do not
see the dollar sign, then the system is not ready for your next input. This applies for the duration
of your session. Later, we will talk about what happens if a command is entered and a $ prompt does
not return to your screen. When typing commands, either from these lessons or from other sources,
do not type the prompt $, only the commands that follow it.
Type the command whoami, then press the enter key (sometimes marked Return) to send the
command to the shell. The command’s output is the ID of the current user, i.e., it shows us who the
shell
thinks we are:
$ whoami
traine
More specifically, when we type whoami the shell:
- finds a program called
whoami, - runs that program,
- displays that program’s output, then
- displays a new prompt to tell us that it’s ready for more commands.
Next, let’s find out where we are by running a command called pwd (which stands for “print working
directory”). At any moment, our current working directory (where we are) is the directory that
the computer assumes we want to run commands in unless we explicitly specify something else. Here,
the computer’s response is /home/1201, which is user
traine’s home directory.
Note that the location of your home directory may differ from system to system.
$ pwd
/home/1201
So, we know where we are. How do we look and see what’s in our current directory?
$ ls
ls prints the names of the files and directories in the current directory in alphabetical order,
arranged neatly into columns.
Differences between remote and local system
Open a second terminal window on your local computer and run the
lscommand without logging in remotely. What differences do you see? Note: Window users will not be able to do this with PuTTY, since PuTTY only opens remote connections.Solution
You would likely see something more like this:
Applications Documents Library Music Public Desktop Downloads Movies PicturesIn addition, you should also note that the text before the prompt (
$) is different. This is very important for making sure you know what system you are issuing commands on when in the shell.
If nothing shows up when you run ls, it means that nothing’s there. Let’s make a directory for us
to play with.
mkdir <new directory name> makes a new directory with that name in your current location. Notice
that this command required two pieces of input: the actual name of the command (mkdir) and an
argument that specifies the name of the directory you wish to create.
$ mkdir training
Let’s type ls again. What do we see?
training
Our folder (directory) is there, awesome. What if we wanted to go inside it and do stuff there? We will use the
cd (change directory) command to move around. Let’s cd into our new training folder.
$ cd training
$ pwd
/home/1201/training
Now that we know how to use cd, we can go anywhere. That’s a lot of responsibility. What happens
if we get “lost” and want to get back to where we started?
To go back to your home directory, the following two commands will work:
$ cd /home/1201
$ cd ~
What is the ~ character? When using the shell, ~ is a shortcut that represents
/home/1201.
A quick note on the structure of a UNIX (Linux/Mac/Android/Solaris/etc) filesystem. Directories and
absolute paths (i.e. exact position in the system) are always prefixed with a /. / is the “root”
or base directory.
Let’s go there now, look around, and then return to our home directory.
$ cd /
$ ls
bin dev home lib64 media opt root sbin sys ttt var
boot etc lib lustre mnt proc run srv tmp usr work
The “home” directory is the one where we generally want to keep all of our files. Other folders on a UNIX OS contain system files, and get modified and changed as you install new software or upgrade your OS.
Returning to your home directory
From the root directory please provide three different ways of getting back to your home directory.
Solution
cd- Shortcut
cd ~- Shorthand
cd /home/1201- This is the absolute path
Using HPC filesystems
On HPC systems, you have a number of places where you can store your files. These differ in both the amount of space allocated and whether they are backed up or not. Caviness has three different types of storage where you can store files that serve different purposes. The amount of storage available and how they are backed up differs as well.
File storage locations:
Home Directory Storage - Each user has 20 GB of disk storage reserved for personal use on the home filesystem. Users’ home directories are in /home (e.g., /home/1201), and the directory name is put in the environment variable
$HOMEat login. The permanent filesystem is configured to allow nearly instantaneous, consistent snapshots. The snapshot contains the original version of the filesystem, and the live filesystem contains any changes made since the snapshot was taken. In addition, all your files are regularly replicated at UD’s off-campus disaster recovery site. You can use read-only snapshots to revert a previous version, or request to have your files restored from the disaster recovery site.You can check to see the size and usage of your home directory with the command
df -h $HOMEWork Group Storage - Each research group has at least 1000 GB of shared group (workgroup) storage in the
/workdirectory identified by the«investing_entity»(e.g.,/work/it_css) and is referred to as your workgroup directory. This is used for input files, supporting data files, work files, and output files, source code and executables that need to be shared with your research group. Just as your home directory, read-only snapshots of workgroup’s files are made several times for the passed week. In addition, the filesystem is replicated on UD’s off-campus disaster recovery site. Snapshots are user-accessible, and older files may be retrieved by special request.You can check the size and usage of your workgroup directory by using the workgroup command to spawn a new workgroup shell, which sets the environment variable
$WORKDIRdf -h $WORKDIRHigh-performance filesystem:
Lustre Storage - User storage is available on a high-performance Lustre-based filesystem having 210 TB of usable space. This is used for temporary input files, supporting data files, work files, and output files associated with computational tasks ran on the cluster. The filesystem is accessible to all of the processor cores via Omni-path Infiniband.
The Lustre filesystem is not backed up nor are there snapshots to recover deleted files. However, it is a robust RAID-6 system. Thus, the filesystem can survive a concurrent disk failure of two independent hard drives and still rebuild its contents automatically.
All users have access the public scratch directory (
/lustre/scratch).A full system inhibits use for everyone potentially preventing jobs from running. Therefore IT staff may run cleanup procedures as needed to purge aged files or directories in Lustre if old files are degrading system performance.
There are several other useful shortcuts you should be aware of.
.represents your current directory..represents the “parent” directory of your current location- While typing nearly anything, you can have bash try to autocomplete what you are typing by
pressing the
tabkey.
Let’s try these out now:
$ cd ./training
$ pwd
$ cd ..
$ pwd
/home/1201/training
/home/1201
Many commands also have multiple behaviours that you can invoke with command line ‘flags.’ What is a
flag? It’s generally just your command followed by a - or -- and the name of the flag.
You follow the flag(s) with any additional arguments you
might need.
We’re going to demonstrate a couple of these “flags” using ls.
Show hidden files with -a. Hidden files are files that begin with ., these files will not appear
otherwise, but that doesn’t mean they aren’t there! “Hidden” files are not hidden for security
purposes, they are usually just config files and other tempfiles that the user doesn’t necessarily
need to see all the time.
$ ls -a
. .. .bash_history .bash_logout .bash_profile .bashrc .bash_udit training .ssh
Notice how both . and .. are visible as hidden files.
To show files, their size in bytes, date last modified, permissions, and other
things with -l.
$ ls -l
drwxr-xr-x 2 traine everyone 2 Jul 13 15:48 training
This is a lot of information to take in at once, but we will explain this later! ls -l is
extremely useful, and tells you almost everything you need to know about your files without
actually looking at them.
We can also use multiple flags at the same time!
$ ls -l -a
[traine@login00 ~]$ ls -la
total 36
drwx--x--x 17 traine everyone 29 Jul 21 14:22 .
drwxr-xr-x 79 root root 0 Jul 21 14:23 ..
-rw------- 1 traine it_css 20442 Jul 21 15:24 .bash_history
-rw-r--r-- 1 traine everyone 17 Jul 25 2018 .bash_logout
-rw-r--r-- 1 traine everyone 200 Jul 25 2018 .bash_profile
-rw-r--r-- 1 traine everyone 384 Mar 19 15:21 .bashrc
-rw-r--r-- 1 traine everyone 1154 Mar 24 10:16 .bash_udit
drwxr-xr-x 2 traine everyone 2 Jul 13 15:48 training
drwx------ 2 traine everyone 6 Mar 18 2020 .ssh
-rw------- 1 traine everyone 1007 Oct 15 11:29 .Xauthority
Flags generally precede any arguments passed to a UNIX command. ls actually takes an extra
argument that specifies a directory to look into. When you use flags and arguments together, they
syntax (how it’s supposed to be typed) generally looks something like this:
$ command <flags/options> <arguments>
So using ls -l -a on a different directory than the one we’re in would look something like:
$ ls -l -a ~/training
drwxr-sr-x 2 yourUsername tc001 4096 Nov 28 09:58 .
drwx--S--- 5 yourUsername tc001 4096 Nov 28 09:58 ..
Another useful flag is the -F flag. The -F appends indicators to entries which helps you
identify files verus directories.
ls -F -la
drwx--x--x 17 traine everyone 29 Jul 21 14:22 ./
drwxr-xr-x 79 root root 0 Jul 21 14:23 ../
-rw------- 1 traine it_css 20442 Jul 21 15:24 .bash_history
-rw-r--r-- 1 traine everyone 17 Jul 25 2018 .bash_logout
-rw-r--r-- 1 traine everyone 200 Jul 25 2018 .bash_profile
-rw-r--r-- 1 traine everyone 384 Mar 19 15:21 .bashrc
-rw-r--r-- 1 traine everyone 1154 Mar 24 10:16 .bash_udit
drwxr-xr-x 2 traine everyone 2 Jul 13 15:48 training/
drwx------ 2 traine everyone 6 Mar 18 2020 .ssh/
-rw------- 1 traine everyone 1007 Oct 15 11:29 .Xauthority
As you can see in the above output rows that end with a / are directories. The rows that don’t
end with a / are files.
Where to go for help?
How did I know about the -l and -a options? Is there a manual we can look at for help when we
need help? There is a very helpful manual for most UNIX commands: man (if you’ve ever heard of a
“man page” for something, this is what it is).
$ man ls
LS(1) User Commands LS(1)
NAME
ls - list directory contents
SYNOPSIS
ls [OPTION]... [FILE]...
DESCRIPTION
List information about the FILEs (the current directory by default). Sort entries alphabetically if none of -cftu‐
vSUX nor --sort is specified.
Mandatory arguments to long options are mandatory for short options too.
Manual page ls(1) line 1 (press h for help or q to quit)
To navigate through the man pages, you may use the up and down arrow keys to move line-by-line, or
try the spacebar and “b” keys to skip up and down by full page. Quit the man pages by typing “q”.
Alternatively, most commands you run will have a --help option that displays addition information
For instance, with ls:
$ ls --help
Usage: ls [OPTION]... [FILE]...
List information about the FILEs (the current directory by default).
Sort entries alphabetically if none of -cftuvSUX nor --sort is specified.
Mandatory arguments to long options are mandatory for short options too.
-a, --all do not ignore entries starting with .
-A, --almost-all do not list implied . and ..
--author with -l, print the author of each file
-b, --escape print C-style escapes for nongraphic characters
--block-size=SIZE scale sizes by SIZE before printing them; e.g.,
'--block-size=M' prints sizes in units of
1,048,576 bytes; see SIZE format below
-B, --ignore-backups do not list implied entries ending with ~
# further output omitted for clarity
Unsupported command-line options
If you try to use an option that is not supported,
lsand other programs will print an error message similar to this:[traine@login00 ~]$ $ ls -jls: invalid option -- 'j' Try 'ls --help' for more information.
Looking at documentation
Looking at the man page for
lsor usingls --help, what does the-h(--human-readable) option do?Solution
-h, --human-readable: with -l, print sizes in human readable format (e.g., 1K 234M 2G)
Absolute vs Relative Paths
Starting from
/Users/amanda/data/, which of the following commands could Amanda use to navigate to her home directory, which is/Users/amanda?
cd .cd /cd /home/amandacd ../..cd ~cd homecd ~/data/..cdcd ..Solution
- No:
.stands for the current directory.- No:
/stands for the root directory.- No: Amanda’s home directory is
/Users/amanda.- No: this goes up two levels, i.e. ends in
/Users.- Yes:
~stands for the user’s home directory, in this case/Users/amanda.- No: this would navigate into a directory
homein the current directory if it exists.- Yes: unnecessarily complicated, but correct.
- Yes: shortcut to go back to the user’s home directory.
- Yes: goes up one level.
Relative Path Resolution
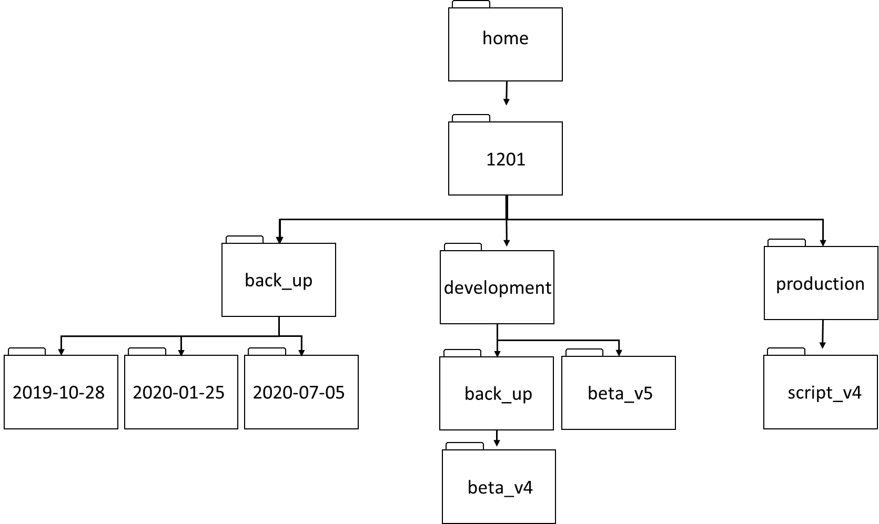
Using the filesystem diagram below, if
pwddisplays/home/1201/development, what willls -F ../back_updisplay?
../back_up: No such file or directorybeta_v4/2019-10-28/ 2020-01-25/ 2020-07-05/beta_v5/ back_up/
Solution
- No: there is a directory
back_upin/home/1201.- No: this is the content of
/home/1201/development/back_up.- Yes: this is the contents of
/home/1201/back_up.- No: this would be the contents of
/home/1201/developmentor./.
lsReading ComprehensionAssuming a directory structure as in the above diagram, if
pwddisplays/home/1201/development, and-rtellslsto display things in reverse order, what command will display:beta_v5/ back_up/
ls pwdls -r -Fls -r -F /home/1201/development- Either #2 or #3 above, but not #1.
Solution
- No:
pwdis not the name of a directory.- Yes:
lswithout directory argument lists files and directories in the current directory.- Yes: uses the absolute path explicitly.
- Correct: see explanations above.
Exploring More
lsArgumentsWhat does the command
lsdo when used with the-land-harguments?Some of its output is about properties that we do not cover in this lesson (such as file permissions and ownership), but the rest should be useful nevertheless.
Solution
The
-larguments makeslsuse a long listing format, showing not only the file/directory names but also additional information such as the file size and the time of its last modification. The-hargument makes the file size “human readable”, i.e. display something like5.3Kinstead of5369.
Listing Recursively and By Time
The command
ls -Rlists the contents of directories recursively, i.e., lists their sub-directories, sub-sub-directories, and so on in alphabetical order at each level. The commandls -tlists things by time of last change, with most recently changed files or directories first. In what order doesls -R -tdisplay things? Hint:ls -luses a long listing format to view timestamps.Solution
The directories are listed alphabetical at each level, the files/directories in each directory are sorted by time of last change.
Key Points
Your current directory is referred to as the working directory.
To change directories, use
cd.To view files, use
ls.You can view help for a command with
man commandorcommand --help.Hit the TAB key to autocomplete whatever you’re currently typing.
Writing and reading files
Overview
Teaching: 30 min
Exercises: 15 minQuestions
How do I create/edit text files?
How do I move/copy/delete files?
Objectives
Learn to use the
nanotext editor.Understand how to move, create, and delete files.
Now that we know how to move around and look at things, let’s learn how to read, write, and handle files! We’ll start by moving back to our home directory and creating a scratch directory:
$ cd ~/training
$ mkdir Documents
Creating and Editing Text Files
When working on an HPC system, we will frequently need to create or edit text files. Text is one of the simplest computer file formats, defined as a simple sequence of text lines.
What if we want to make a file? There are a few ways of doing this, the easiest of which is simply
using a text editor. For this lesson, we are going to us nano, since it’s more intuitive than many
other terminal text editors.
To create or edit a file, type nano <filename>, on the terminal, where <filename> is the name of the
file. If the file does not already exist, it will be created.
Let’s make a new file now, type whatever you want in it, and save it.
$ nano draft.txt
Add the following text It's not "publish or perish" anymore, it's "share and thrive.
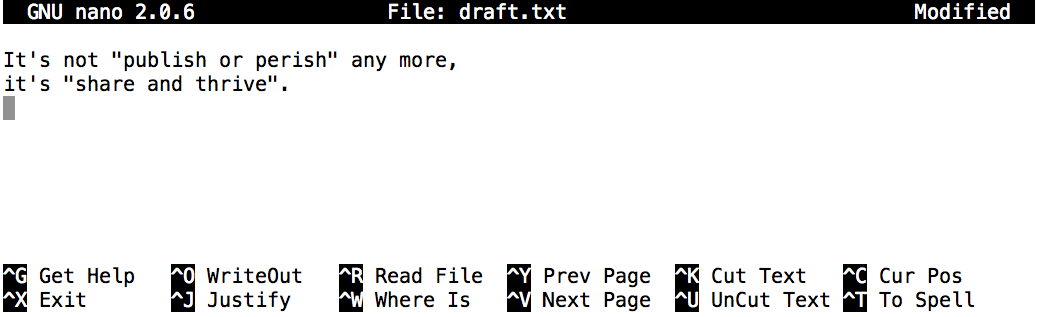
Nano defines a number of shortcut keys (prefixed by the Control or Ctrl key) to perform actions such as saving the file or exiting the editor. Here are the shortcut keys for a few common actions:
-
Ctrl+O — save the file (into a current name or a new name).
-
Ctrl+X — exit the editor. If you have not saved your file upon exiting,
nanowill ask you if you want to save. -
Ctrl+K — cut (“kill”) a text line. This command deletes a line and saves it on a clipboard. If repeated multiple times without any interruption (key typing or cursor movement), it will cut a chunk of text lines.
-
Ctrl+U — paste the cut text line (or lines). This command can be repeated to paste the same text elsewhere.
Now save your file and close nano.
Using
vimas a text editorFrom time to time, you may encounter the
vimtext editor. Althoughvimisn’t the easiest or most user-friendly of text editors, you’ll be able to find it on any system and it has many more features thannano.
vimhas several modes, a “command” mode (for doing big operations, like saving and quitting) and an “insert” mode. You can switch to insert mode with theikey, and command mode withEsc.In insert mode, you can type more or less normally. In command mode there are a few commands you should be aware of:
:q!— quit, without saving:wq— save and quitdd— cut/delete a liney— paste a line
Do a quick check to confirm our file was created.
$ ls
Documents draft.txt
Another way to create a file is with the touch command. The touch command is a simple way to
create a empty file or to update the modified time of a file that already exists.
$ touch draft2.txt
$ ls
Documents draft2.txt draft.txt
Reading Files
Let’s read the file we just created now. There are a few different ways of doing this, one of which is
reading the entire file with cat.
$ cat draft.txt
It's not "publish or perish" any more,
it's "share and thrive".
By default, cat prints out the content of the given file.
Although cat may not seem like an intuitive command with which to read files, it stands for
“concatenate”. Giving it multiple file names will print out the contents of the input files in the order
specified in the cat’s invocation.
For example,
$ cat draft.txt draft.txt
It's not "publish or perish" any more,
it's "share and thrive".
It's not "publish or perish" any more,
it's "share and thrive".
Reading Multiple Text Files
Create two more files using
nano, giving them different names such aschap1.txtandchap2.txt. Add unique text into them such as “This is chap1.txt” and “This is chap2.txt” repectively. Then use a singlecatcommand to read and print the contents ofdraft.txt,chap1.txt, andchap2.txt.Solution
$ cat draft.txt chap1.txt chap2.txt It's not "publish or perish" any more, it's "share and thrive". This is chap1.txt This is chap2.txt
Creating Directory
We’ve successfully created a file. What about a directory? We’ve actually done this before, using
mkdir.
$ mkdir files
$ ls
Documents draft2.txt draft.txt files
Moving, Renaming, Copying Files
Moving—We will move draft.txt to the files directory with mv (“move”) command.
The same syntax works for both files and directories: mv <file/directory> <new-location>
$ mv draft.txt files
$ cd files
$ ls
draft.txt
Renaming—draft.txt isn’t a very descriptive name. How do we go about changing it?
It turns out that mv is also used to rename files and directories. Although this may not seem
intuitive at first, think of it as moving a file to be stored under a different name. The syntax is
quite similar to moving files: mv oldName newName.
$ mv draft.txt newname.testfile
$ ls
newname.testfile
File extensions are arbitrary
In the last example, we changed both a file’s name and extension at the same time. On UNIX systems, file extensions (like
.txt) are arbitrary. A file is a.txtfile only because we say it is. Changing the name or extension of the file will never change a file’s contents, so you are free to rename things as you wish. With that in mind, however, file extensions are a useful tool for keeping track of what type of data it contains. A.txtfile typically contains text, for instance. The extensions may also be used by other applications to help interpret the files contents. For example changing.mp3to.txtmight make the file unreadable to a music player.
Copying—What if we want to copy a file, instead of simply renaming or moving it?
Use cp command (an abbreviated name for “copy”). This command has two different uses that work in the same way as mv:
- Copy to same directory (copied file is renamed):
cp file newFilename - Copy to other directory (copied file retains original name):
cp file directory
Let’s try this out.
$ cp newname.testfile copy.testfile
$ ls
$ cp newname.testfile ..
$ cd ..
$ ls
newname.testfile copy.testfile
Documents draft2.txt files newname.testfile
Removing files
Warning About Removing Files
We’ve begun to clutter up our workspace with all of the directories and files we’ve been making. Let’s learn how to get rid of them. One important note before we start… when you delete a file on UNIX systems, they are gone forever. There is no “recycle bin” or “trash”. Once a file is deleted, it is gone, never to return. So be very careful when deleting files.
Files are deleted with rm file [moreFiles]. To delete the newname.testfile in our current
directory:
$ ls
$ rm newname.testfile
$ ls
Documents draft2.txt files newname.testfile
Documents draft2.txt files
That was simple enough. Directories are deleted in a similar manner using rm -r (the -r option
stands for ‘recursive’).
$ ls
$ rm Documents
$ rmdir Documents
$ rmdir files
$ rm -r files
$ ls
files Documents
rm: cannot remove ‘Documents’: Is a directory
rmdir: failed to remove `files/': Directory not empty
draft2.txt
What happened? As it turns out, rmdir is unable to remove directories that have stuff in them. To
delete a directory and everything inside it, we will use a special variant of rm, rm -r
directory. This is probably the scariest command on UNIX- it will force delete a directory and all
of its contents without prompting. ALWAYS double check your typing before using it… if you
leave out the arguments, it will attempt to delete everything on your file system that you have
permission to delete. So when deleting directories be very, very careful.
What happens when you use
rm -rfaccidentallySteam is a major online sales platform for PC video games with over 125 million users. Despite this, it hasn’t always had the most stable or error-free code.
In January 2015, user kevyin on GitHub reported that Steam’s Linux client had deleted every file on his computer. It turned out that one of the Steam programmers had added the following line:
rm -rf "$STEAMROOT/"*. Due to the way that Steam was set up, the variable$STEAMROOTwas never initialized, meaning the statement evaluated torm -rf /*. This coding error in the Linux client meant that Steam deleted every single file on a computer when run in certain scenarios (including connected external hard drives). Moral of the story: be very careful when usingrm -rf!
Now before moving on lets finish cleaning up the files and directories that we created and will no longer be need.
$ cd ~
$ ls
$ rm -r Documents
$ls -a
Documents
. ..
Looking at files
Sometimes it’s not practical to read an entire file with cat the file might be way too large,
take a long time to open, or maybe we want to only look at a certain part of the file. As an
example, we are going to look at a large and complex file type used in bioinformatics, .gtf file extension.
The GTF2 format is commonly used to describe the location of genetic features in a genome.
Let’s grab and unpack a set of demo files for use later. To do this, we’ll use wget (wget url
downloads a file from a url).
$ cd ~
$ mkdir lesson_files
$ cd lesson_files
$ wget https://mkyleud.github.io/Caviness-HPC-Shell/files/bash-lesson.tar.gz
Problems with
wget?
wgetis a stand-alone application for downloading things over HTTP/HTTPS and FTP/FTPS connections, and it does the job admirably – when it is installed.Some operating systems instead come with cURL, which is the command-line interface to
libcurl, a powerful library for programming interactions with remote resources over a wide variety of network protocols. If you havecurlbut notwget, then try this command instead:$ curl -O https://mkyleud.github.io/Caviness-HPC-Shell/files/bash-lesson.tar.gzFor very large downloads, you might consider using Aria2, which has support for downloading the same file from multiple mirrors. You have to install it separately, but if you have it, try this to get it faster than your neighbors:
$ aria2c https://mkyleud.github.io/Caviness-HPC-Shell/files/bash-lesson.tar.gz https://hpc-carpentry.github.io/hpc-shell/files/bash-lesson.tar.gzInstall cURL
- macOS:
curlis pre-installed on macOS. If you must have the latest version you canbrew installit, but only do so if the stock version has failed you.Windows:
curlcomes preinstalled for the Windows 10 command line. For earlier Windows systems, you can download the executable directly; run it in place.
curlcomes preinstalled in Git for Windows and Windows Subsystem for Linux. On Cygwin, run the setup program again and select thecurlpackage to install it.- Linux:
curlis packaged for every major distribution. You can install it through the usual means.
- Debian, Ubuntu, Mint:
sudo apt install curl- CentOS, Red Hat:
sudo yum install curlorzypper install curl- Fedora:
sudo dnf install curlInstall Aria2
- macOS:
aria2cis available through a homebrew.brew install aria2.- Windows: download the latest release and run
aria2cin place. If you’re using the Windows Subsystem for Linux,- Linux: every major distribution has an
aria2package. Install it by the usual means.
- Debian, Ubuntu, Mint:
sudo apt install aria2- CentOS, Red Hat:
sudo yum install aria2orzypper install aria2- Fedora:
sudo dnf install aria2
You’ll commonly encounter .tar.gz archives while working in UNIX. To extract the files from a
.tar.gz file, we run the command tar -xvf filename.tar.gz:
$ tar -xvf bash-lesson.tar.gz
dmel-all-r6.19.gtf
dmel_unique_protein_isoforms_fb_2016_01.tsv
gene_association.fb
SRR307023_1.fastq
SRR307023_2.fastq
SRR307024_1.fastq
SRR307024_2.fastq
SRR307025_1.fastq
SRR307025_2.fastq
SRR307026_1.fastq
SRR307026_2.fastq
SRR307027_1.fastq
SRR307027_2.fastq
SRR307028_1.fastq
SRR307028_2.fastq
SRR307029_1.fastq
SRR307029_2.fastq
SRR307030_1.fastq
SRR307030_2.fastq
Unzipping files
We just unzipped a .tar.gz file for this example. What if we run into other file formats that we need to unzip? Just use the handy reference below:
gunzipextracts the contents of.gzfilesunzipextracts the contents of.zipfilestar -xvfextracts the contents of.tar.gzand.tar.bz2files
That is a lot of files! One of these files, dmel-all-r6.19.gtf is extremely large, and contains
every annotated feature in the Drosophila melanogaster genome. It’s a huge file- what happens if
we run cat on it? (Press Ctrl + C to stop it).
So, cat is a really bad option when reading big files… it scrolls through the entire file far
too quickly! What are the alternatives? Try all of these out and see which ones you like best!
head file: Print the top 10 lines in a file to the console. You can control the number of lines you see with the-n numberOfLinesflag.tail file: Same ashead, but prints the last 10 lines in a file to the console.less file: Opens a file and display as much as possible on-screen. You can scroll withEnteror the up and down arrow keys on your keyboard. Pressqto close the viewer.more file: Opens a file and display as much as possible on-screen. You can scroll withEnterbut not the arrow keys on your keyboard. Pressqto close the viewer.
Out of cat, head, tail, less, and more, which method of reading files is your favorite? Why?
Key Points
Use
nanoto create or edit text files from a terminal.
cat file1 [file2 ...]prints the contents of one or more files to terminal.
mv old dirmoves a file or directory to another directorydir.
mv old newrenames a file or directory.
cp old newcopies a file.
cp old dircopies a file to another directorydir.
rm pathdeletes (removes) a file.File extensions are entirely arbitrary on UNIX systems. However they are important for applications to recognize files they use.
Wildcards and pipes
Overview
Teaching: 45 min
Exercises: 10 minQuestions
How can I run a command on multiple files at once?
Is there an easy way of saving a command’s output?
Objectives
Redirect a command’s output to a file.
Process a file instead of keyboard input using redirection.
Construct command pipelines with two or more stages.
Explain what usually happens if a program or pipeline isn’t given any input to process.
Required files
If you didn’t get them in the last lesson, make sure to download the example files used in the next few sections:
Using wget —
wget https://mkyleud.github.io/Caviness-HPC-Shell/files/bash-lesson.tar.gz$ cd ~ $ mkdir lesson_files $ cd lesson_files $ wget https://mkyleud.github.io/Caviness-HPC-Shell/files/bash-lesson.tar.gz $ tar -xvf bash-lesson.tar.gz
Now that we know some of the basic UNIX commands, we are going to explore some more advanced
features. The first of these features is the wildcard *. In our examples before, we’ve done things
to files one at a time and otherwise had to specify things explicitly. The * character lets us
speed things up and do things across multiple files.
Ever wanted to move, delete, or just do “something” to all files of a certain type in a directory?
* lets you do that, by taking the place of zero or more characters in a piece of text. So *.txt
would be equivalent to all .txt files in a directory for instance. * by itself means all files.
Let’s use our example data to see what I mean.
$ ls
bash-lesson.tar.gz SRR307024_2.fastq SRR307028_1.fastq
dmel-all-r6.19.gtf SRR307025_1.fastq SRR307028_2.fastq
dmel_unique_protein_isoforms_fb_2016_01.tsv SRR307025_2.fastq SRR307029_1.fastq
gene_association.fb SRR307026_1.fastq SRR307029_2.fastq
SRR307023_1.fastq SRR307026_2.fastq SRR307030_1.fastq
SRR307023_2.fastq SRR307027_1.fastq SRR307030_2.fastq
SRR307024_1.fastq SRR307027_2.fastq
Now we have a whole bunch of example files in our directory. For this example we are going to learn
a new command that tells us how long a file is: wc. wc -l file tells us the length of a file in
lines.
$ wc -l dmel-all-r6.19.gtf
542048 dmel-all-r6.19.gtf
Interesting, there are over 540000 lines in our dmel-all-r6.19.gtf file. What if we wanted to run
wc -l on every .fastq file? This is where * comes in really handy! *.fastq would match every
file ending in .fastq.
$ wc -l *.fastq
20000 SRR307023_1.fastq
20000 SRR307023_2.fastq
20000 SRR307024_1.fastq
20000 SRR307024_2.fastq
20000 SRR307025_1.fastq
20000 SRR307025_2.fastq
20000 SRR307026_1.fastq
20000 SRR307026_2.fastq
20000 SRR307027_1.fastq
20000 SRR307027_2.fastq
20000 SRR307028_1.fastq
20000 SRR307028_2.fastq
20000 SRR307029_1.fastq
20000 SRR307029_2.fastq
20000 SRR307030_1.fastq
20000 SRR307030_2.fastq
320000 total
That was easy. What if we wanted to do the same command, except on every file in the directory? A
nice trick to keep in mind is that * by itself matches every file.
$ wc -l *
53037 bash-lesson.tar.gz
542048 dmel-all-r6.19.gtf
22129 dmel_unique_protein_isoforms_fb_2016_01.tsv
106290 gene_association.fb
20000 SRR307023_1.fastq
20000 SRR307023_2.fastq
20000 SRR307024_1.fastq
20000 SRR307024_2.fastq
20000 SRR307025_1.fastq
20000 SRR307025_2.fastq
20000 SRR307026_1.fastq
20000 SRR307026_2.fastq
20000 SRR307027_1.fastq
20000 SRR307027_2.fastq
20000 SRR307028_1.fastq
20000 SRR307028_2.fastq
20000 SRR307029_1.fastq
20000 SRR307029_2.fastq
20000 SRR307030_1.fastq
20000 SRR307030_2.fastq
1043504 total
Multiple wildcards
You can even use multiple
*s at a time. How would you runwc -lon every file with “fb” in it?Solution
wc -l *fb*i.e. anything or nothing then
fbthen anything or nothing
Using other commands
Now let’s try cleaning up our working directory a bit. Create a folder called “fastq” and move all of our .fastq files there in one
mvcommand.Solution
mkdir fastq mv *.fastq fastq/
Redirecting output
Each of the commands we’ve used so far does only a very small amount of work. However, we can chain these small UNIX commands together to perform otherwise complicated actions!
For our first foray into redirecting output, we are going to use the > operator to
write output to a file. When using >, whatever is on the left of the > is written to the
filename you specify on the right of the arrow. The actual syntax looks like command > filename.
Let’s try several basic usages of >. echo simply prints back, or echoes whatever you type after
it.
$ echo "this is a test"
this is a test
$
$ echo "this is a test" > test.txt
# This is empty because the output was directed into the test.txt file.
$ ls
bash-lesson.tar.gz fastq
dmel-all-r6.19.gtf gene_association.fb
dmel_unique_protein_isoforms_fb_2016_01.tsv test.txt
$ cat test.txt
this is a test
Awesome, let’s try that with a more complicated command, like wc -l.
$ wc -l * > word_counts.txt
wc: fastq: Is a directory
$ cat word_counts.txt
53037 bash-lesson.tar.gz
542048 dmel-all-r6.19.gtf
22129 dmel_unique_protein_isoforms_fb_2016_01.tsv
0 fastq
106290 gene_association.fb
1 test.txt
723505 total
Notice how we still got some output to the console even though we “redirected” the output to a file? Our expected output still went to the file, but how did the error message get skipped and not go to the file?
This phenomena is an artefact of how UNIX systems are built. There are 3 input/output streams for
every UNIX program you will run: stdin, stdout, and stderr.
Let’s dissect these three streams of input/output in the command we just ran: wc -l * >
word_counts.txt
-
stdinis the input to a program. In the command we just ran,stdinis represented by*, which is simply every filename in our current directory. -
stdoutcontains the actual, expected output. In this case,>redirectedstdoutto the fileword_counts.txt. -
stderrtypically contains error messages and other information that doesn’t quite fit into the category of “output”. If we insist on redirecting bothstdoutandstderrto the same file we would use&>instead of>. (We can redirect juststderrusing2>.)
Knowing what we know now, let’s try re-running the command, and send all of the output (including
the error message) to the same word_counts.txt files as before.
$ wc -l * &> word_counts.txt
Notice how there was no output to the console that time. Let’s check that the error message went to the file like we specified.
$ cat word_counts.txt
53037 bash-lesson.tar.gz
542048 dmel-all-r6.19.gtf
22129 dmel_unique_protein_isoforms_fb_2016_01.tsv
wc: fastq: Is a directory
0 fastq
106290 gene_association.fb
1 test.txt
7 word_counts.txt
723512 total
Success! The wc: fastq: Is a directory error message was written to the file. Also, note how the
file was silently overwritten by directing output to the same place as before. Sometimes this is not
the behavior we want. How do we append (add) to a file instead of overwriting it?
Appending to a file is done the same was as redirecting output. However, instead of >, we will use
>>.
$ echo "We want to add this sentence to the end of our file" >> word_counts.txt
$ cat word_counts.txt
22129 dmel_unique_protein_isoforms_fb_2016_01.tsv
471308 Drosophila_melanogaster.BDGP5.77.gtf
0 fastq
1304914 fb_synonym_fb_2016_01.tsv
106290 gene_association.fb
1 test.txt
1904642 total
We want to add this sentence to the end of our file
Chaining commands together
We now know how to redirect stdout and stderr to files. We can actually take this a step further
and redirect output (stdout) from one command to serve as the input (stdin) for the next. This is
referred to as piping by using the | (pipe) operator.
grep is an extremely useful command. It finds things for us within files. Basic usage (there are a
lot of options for more clever things, see the man page) uses the syntax grep whatToFind
fileToSearch. Let’s use grep to find all of the entries pertaining to the Act5C gene in
Drosophila melanogaster.
$ grep Act5C dmel-all-r6.19.gtf
The output is nearly unintelligible since there is so much of it. Let’s send the output of that
grep command to head so we can just take a peek at the first line. The | operator lets us send
output from one command to the next:
$ grep Act5C dmel-all-r6.19.gtf | head -n 1
X FlyBase gene 5900861 5905399 . + . gene_id "FBgn0000042"; gene_symbol "Act5C";
Nice work, we sent the output of grep to head. Let’s try counting the number of entries for
Act5C with wc -l. We can do the same trick to send grep’s output to wc -l:
$ grep Act5C dmel-all-r6.19.gtf | wc -l
46
Note that this is just the same as redirecting output to a file, then reading the number of lines from that file.
Writing commands using pipes
What command will tell how many files are there in the “fastq” directory? How many are there? (Use the shell to do this.)
Solution
ls fastq/ | wc -l16Output of
lsis one line per item so counting lines gives the number of files.
Reading from compressed files
Let’s compress one of our files using gzip.
$ gzip gene_association.fb
zcatacts likecat, except that it can read information from.gz(compressed) files. Usingzcat, can you write a command to take a look at the top few lines of thegene_association.fb.gzfile (without decompressing the file itself)?Solution
zcat gene_association.fb.gz | headThe
headcommand without any options shows the first 10 lines of a file
Key Points
The
*wildcard is used as a placeholder to match any text that follows a pattern.Redirect a commands output to a file with
>.Commands can be chained with
|
Scripts, variables, and loops
Overview
Teaching: 45 min
Exercises: 10 minQuestions
How do I turn a set of commands into a program?
Objectives
Write a shell script
Understand and manipulate UNIX permissions
Understand shell variables and how to use them
Write a simple
forloop
We now know a lot of UNIX commands! Wouldn’t it be great if we could save certain commands so that we could run them later or not have to type them out again? As it turns out, this is straightforward to do. A “shell script” is essentially a text file containing a list of UNIX commands to be executed in a sequential manner. These shell scripts can be run whenever we want, and are a great way to automate our work. Before we get started lets make sure that we are all in the same directory
$ cd ~/lesson_files
$ pwd
/home/1201/lesson_files
Keep An Eye On Spacing…
When creating a new variable or working on a script, it is very important to monitor when to use, and when not to use a space—spacing is very important. Using a space in the wrong spot can have consequences that will cause your code to not work correctly.
Writing a Script
So how do we write a shell script, exactly? It turns out we can do this with a text editor.
Start editing a file called “demo.sh” (to recap, we can do this with nano demo.sh). The “.sh” is
the standard file extension for shell scripts that most people use (you may also see “.bash” used).
Our shell script will have two parts:
- On the very first line, add
#!/bin/bash. The#!(pronounced “hash-bang”) tells our computer what program to run our script with. In this case, we are telling it to run our script with our command-line shell (what we’ve been doing everything in so far). If we wanted our script to be run with something else, like Perl, we could add#!/usr/bin/perl - Now, anywhere below the first line, add
echo "Our script worked!". When our script runs,echowill happily print outOur script worked!.
Our file should now look like this:
#!/bin/bash
echo "Our script worked!"
Ready to run our program? Let’s try running it:
$ demo.sh
bash: demo.sh: command not found...
Strangely enough, Bash can’t find our script. As it turns out, Bash will only look in certain
directories for scripts to run. To run anything else, we need to tell Bash exactly where to look. To
run a script that we wrote ourselves, we need to specify the full path to the file, followed by the
filename. We could do this one of two ways: either with our absolute path
/home/1201/lesson_files/demo.sh, or with the relative path ./demo.sh.
$ ./demo.sh
bash: ./demo.sh: Permission denied
There’s one last thing we need to do. Before a file can be run, it needs “permission” to run. Let’s
look at our file’s permissions with ls -l:
$ ls -l
-rw-rw-r--. 1 traine everyone 12534006 Jan 16 18:50 bash-lesson.tar.gz
-rw-rw-r--. 1 traine everyone 40 Jan 16 19:41 demo.sh
-rw-rw-r--. 1 traine everyone 77426528 Jan 16 18:50 dmel-all-r6.19.gtf
-rw-r--r--. 1 traine everyone 721242 Jan 25 2016 dmel_unique_protein_isoforms_fb_2016_01.tsv
drwxrwxr-x. 2 traine everyone 4096 Jan 16 19:16 fastq
-rw-r--r--. 1 traine everyone 1830516 Jan 25 2016 gene_association.fb.gz
-rw-rw-r--. 1 traine everyone 15 Jan 16 19:17 test.txt
-rw-rw-r--. 1 traine everyone 245 Jan 16 19:24 word_counts.txt
That’s a huge amount of output. Let’s see if we can understand what it is, working left to right.
- 1st column - Permissions: On the very left side, there is a string of the characters
d,r,w,x, and-. Thedindicates if something is a directory (there is a-in that spot if it is not a directory). The otherr,w,xbits indicates permission to Read Write and eXecute a file. There are three columns ofrwxpermissions following the spot ford. If a user is missing a permission to do something, it’s indicated by a-.- The first column of
rwxare the permissions that the owner has (in this case the owner istraine). - The second set of
rwxs are permissions that other members of the owner’s group share (in this case, the group is namedtc001). - The third set of
rwxs are permissions that anyone else with access to this computer can do with a file. Though files are typically created with read permissions for everyone, typically the permissions on your home directory prevent others from being able to access the file in the first place.
- The first column of
-
2nd column - Owner: This is the username of the user who owns the file. Their permissions are indicated in the first permissions column.
-
3rd column - Group: This is the user group of the user who owns the file. Members of this user group have permissions indicated in the second permissions column.
-
4th column - Size of file: This is the size of a file in bytes, or the number of files/subdirectories if we are looking at a directory. (We can use the
-hoption here to get a human-readable file size in megabytes, gigabytes, etc.) -
5th column - Time last modified: This is the last time the file was modified.
- 6th column - Filename: This is the filename.
So how do we change permissions? As I mentioned earlier, we need permission to execute our script.
Changing permissions is done with chmod. To add executable permissions for all users we could use
this:
$ chmod +x demo.sh
$ ls -l
-rw-rw-r--. 1 traine everyone 12534006 Jan 16 18:50 bash-lesson.tar.gz
-rwxrwxr-x. 1 traine everyone 40 Jan 16 19:41 demo.sh
-rw-rw-r--. 1 traine everyone 77426528 Jan 16 18:50 dmel-all-r6.19.gtf
-rw-r--r--. 1 traine everyone 721242 Jan 25 2016 dmel_unique_protein_isoforms_fb_2016_01.tsv
drwxrwxr-x. 2 traine everyone 4096 Jan 16 19:16 fastq
-rw-r--r--. 1 traine everyone 1830516 Jan 25 2016 gene_association.fb.gz
-rw-rw-r--. 1 traine everyone 15 Jan 16 19:17 test.txt
-rw-rw-r--. 1 traine everyone 245 Jan 16 19:24 word_counts.txt
Now that we have executable permissions for that file, we can run it.
$ ./demo.sh
Our script worked!
Fantastic, we’ve written our first program! Before we go any further, let’s learn how to take notes
inside our program using comments. A comment is indicated by the # character, followed by whatever
we want. Comments do not get run. Let’s try out some comments in the console, then add one to our
script!
# This wont show anything
Now lets try adding this to our script with nano. Edit your script to look something like this:
#!/bin/bash
# This is a comment... they are nice for making notes!
echo "Our script worked!"
When we run our script, the output should be unchanged from before!
Shell variables
One important concept that we’ll need to cover are shell variables. Variables are a great way of saving information under a name you can access later. In programming languages like Python and R, variables can store pretty much anything you can think of. In the shell, they usually just store text. The best way to understand how they work is to see them in action.
To set a variable, simply type in a name containing only letters, numbers, and underscores, followed
by an = and whatever you want to put in the variable. Shell variable names are often uppercase
by convention (but do not have to be).
$ foo="This is our variable"
To use a variable, prefix its name with a $ sign. Note that if we want to simply check what a
variable is, we should use echo (or else the shell will try to run the contents of a variable).
$ echo $foo
This is our variable
Let’s try setting a variable in our script and then recalling its value as part of a command. We’re
going to make it so our script runs wc -l on whichever file we specify with fileName.
Now lets create a new script called demo1.sh.
$ nano demo1.sh
Our script:
#!/bin/bash
# set our variable to the name of our GTF file
fileName=dmel-all-r6.19.gtf
# call wc -l on our file
wc -l $fileName
Don’t forget to add the execuation permission to the file before trying to run it.
$ chmod +x demo1.sh
$ ./demo1.sh
542048 dmel-all-r6.19.gtf
What if we wanted to do our little wc -l script on other files without having to change $fileName
every time we want to use it? There is actually a special shell variable we can use in scripts that
allows us to use arguments in our scripts (arguments are extra information that we can pass to our
script, like the -l in wc -l).
To use the first argument to a script, use $1 (the second argument is $2, and so on).
Let’s change our script to run wc -l on $1 instead of $fileName. Note that we can also pass all of
the arguments using $@ (not going to use it in this lesson, but it’s something to be aware of).
Now we will create our third script and this time call it demo2.sh.
Our script:
#!/bin/bash
# call wc -l on our first argument
wc -l $1
$ chmod +x demo2.sh
$ ./demo2.sh dmel_unique_protein_isoforms_fb_2016_01.tsv
22129 dmel_unique_protein_isoforms_fb_2016_01.tsv
Nice! One thing to be aware of when using variables: they are all treated as pure text. How do we
save the output of an actual command like ls -l?
A demonstration of what doesn’t work:
$ listVar=ls -l
-bash: -l: command not found
What does work (we need to surround any command with $(command)):
$ listVar=$(ls -l)
$ echo $listVar
total 90372 -rw-rw-r--. 1 jeff jeff 12534006 Jan 16 18:50 bash-lesson.tar.gz -rwxrwxr-x. 1 jeff jeff 40 Jan 1619:41 demo.sh -rw-rw-r--. 1 jeff jeff 77426528 Jan 16 18:50 dmel-all-r6.19.gtf -rw-r--r--. 1 jeff jeff 721242 Jan 25 2016 dmel_unique_protein_isoforms_fb_2016_01.tsv drwxrwxr-x. 2 jeff jeff 4096 Jan 16 19:16 fastq -rw-r--r--. 1 jeff jeff 1830516 Jan 25 2016 gene_association.fb.gz -rw-rw-r--. 1 jeff jeff 15 Jan 16 19:17 test.txt -rw-rw-r--. 1 jeff jeff 245 Jan 16 19:24 word_counts.txt
Note that everything got printed on the same line. This is a feature, not a bug, as it allows us to
use $(commands) inside lines of script without triggering line breaks (which would end our line of
code and execute it prematurely).
Loops
To end our lesson on scripts, we are going to learn how to write a for-loop to execute a lot of commands at once. This will let us do the same string of commands on every file in a directory (or other stuff of that nature).
Now lets create our fourth script called demo3.sh.
$ nano demo3.sh
for-loops generally have the following syntax:
#!/bin/bash
for count in first second third
do
echo $count
done
When a for-loop gets run, the loop will run once for everything following the word in. In each
iteration, the variable $count is set to a particular value for that iteration. In this case it will
be set to first during the first iteration, second on the second, and so on. During each
iteration, the code between do and done is performed.
Let’s run the script we just wrote.
$ chmod +x demo3.sh
$ ./demo3.sh
first
second
third
What if we wanted to loop over a shell variable, such as every file in the current directory? Shell
variables work perfectly in for-loops. In this example, we’ll save the result of ls and loop over
each file.
Now lets create our fifth script called demo4.sh.
$ nano demo4.sh
#!/bin/bash
fileList=$(ls)
for file in $fileList
do
echo $file
done
$ chmod +x demo4.sh
$ ./demo4.sh
bash-lesson.tar.gz
demo1.sh
demo2.sh
demo3.sh
demo4.sh
demo.sh
dmel_unique_protein_isoforms_fb_2016_01.tsv
dmel-all-r6.19.gtf
fastq
gene_association.fb.gz
test.txt
word_counts.txt
There’s actually even a shortcut to run on all files of a particular type, say all .gz files:
#!/bin/bash
for file in *.gz
do
echo $file
done
bash-lesson.tar.gz
gene_association.fb.gz
To tie this all together let’s use what we have learned from the above example and use it in a script
that will make a fastq_bak directory, and then copy the files from the fastq directory, and
rename to the same name, but with “_bak” added to the end of the file name.
This will make use of not just writing a script but also a lot of the other commands that have been covered through out this workshop.
To write this script start by creating a new file with nano called backup_copy.sh. Then add and
review the below code.
#!/bin/bash
## In this script we will create a new directory called fastq_bak.
## Then we will store all the files in the fastq directory to a variable.
## Next we will loop through the variables modify it's name and save it
## into the new directory that was made in the first step.
### Step 1: Make the directory
mkdir fastq_bak
### Step2: Create a list of the files in fastq and setting it the fastqFiles
### variable. Be mindful of the where the fastq files are located with
### regards to your current working directory.
fastqFiles=$(ls fastq)
### Step3: Looping through the files in the fastqFile variable and temporary
### storing each file name in the loop variable file.
for file in $fastqFiles
do
#echo ${file} #can be used for debugging
### Step 4: Modifing the original file name and appendin "_bak" it's end
newName=${file}"_bak"
#echo $newName #can be used for debugging
### Step 5: Copying the file to the new directory with its new name. Again
### keep in mind your current working directory and where you want
### the files to be copied from and too.
cp fastq/${file} fastq_bak/${newName}
done
### Step 6: A quick simple check to see if everything worked.
orgFileCount=$(ls fastq | wc -l) #Getting the file count of the fastq directory
bakFileCount=$(ls fastq_bak | wc -l) #Getting the file count of the fastq_bak directory
#Use an if statement to check if the two counts are equal
if [ ${orgFileCount} -eq ${bakFileCount} ]
then
echo "All ${bakFileCount} files copied successfully"
else
echo "${bakFileCount} of the ${orgFileCount} have been copied to fastq_bak"
fi
Now lets fun this the backup_copy.sh script. Again since this is a new script don’take
forget that we first need to make the file executable.
$ chmod +x backup_copy.sh
$ ./backup_copy.sh
All 16 files copied successfully
Writing our own scripts and loops
cdto ourfastqdirectory from earlier and write a loop to print off the name and top 4 lines of every fastq file in that directory.Is there a way to only run the loop on fastq files ending in
_1.fastq?Solution
Create the following script in a file called
head_all.sh#!/bin/bash for FILE in *.fastq do echo $FILE head -n 4 $FILE doneThe “for” line could be modified to be
for FILE in *_1.fastqto achieve the second aim
Concatenating variables
Concatenating (i.e. mashing together) variables is quite easy to do. Add whatever you want to concatenate to the beginning or end of the shell variable after enclosing it in
{}characters.FILE=stuff.txt echo ${FILE}.examplestuff.txt.exampleCan you write a script that prints off the name of every file in a directory with “.processed” added to it?
Solution
Create the following script in a file called
process.sh#!/bin/bash for FILE in * do echo ${FILE}.processed doneNote that this will also print directories appended with “.processed”. To truely only get files and not directories, we need to modify this to use the
findcommand to give us only files in the current directory:#!/bin/bash for FILE in $(find . -max-depth 1 -type f) do echo ${FILE}.processed donebut this will have the side-effect of listing hidden files too.
Special permissions
What if we want to give different sets of users different permissions.
chmodactually accepts special numeric codes instead of stuff likechmod +x. The numeric codes are as follows: read = 4, write = 2, execute = 1. For each user we will assign permissions based on the sum of these permissions (must be between 7 and 0).Let’s make an example file and give everyone permission to do everything with it.
touch example ls -l example chmod 777 example ls -l exampleHow might we give ourselves permission to do everything with a file, but allow no one else to do anything with it.
Solution
chmod 700 exampleWe want all permissions so: 4 (read) + 2 (write) + 1 (execute) = 7 for user (first position), no permissions, i.e. 0, for group (second position) and all (third position).
Key Points
A shell script is just a list of bash commands in a text file.
chmod +x script.shwill give it permission to execute.